NGSPipes repository¶
The NGSPipes repository is a component of NGSPipes system that contains all the information related to the available tools which can be used when defining a pipeline. We provide a repository prototype that contains some tools to test our system, which can be found in https://github.com/ngspipes/tools. User made repositories can be used, as it will be explained in this section. This component has to supply the following information:
- a list of tool names;
- a list of tool descriptors;
- a list of tool logotypes (optional);
- a list of configurators of a given tool;
- a list of the names of the configurators available for a given tool.
For defining the tool descriptors and configurators, we have defined JSON schemas, as well as for specify all the tools that are available in the repository.
Tool names¶
The repository is composed by a list of tools. All the tools names that are available in a given repository, are described in a file with a JSON format designed by Tools.json.
In NGSPipes repository example this file appears at the root of the repository ( please, see the tool’s repository). Moreover, the presented repository structure is one of the possible structures that is supported by the repository support library used in the NGSPIpes framework. The format of the Tools.json file is given by the JSON schema presented in Listing 3.1.
{
"$schema": "http://json-schema.org/draft-04/schema#",
"type": "object",
"properties": {
"toolsName": {
"type": "array",
"items":{ "type": "string" }
},
"required": [ "toolsName" ]
}
}
Listing 3.1: JSON schema for specifying the names of the tools included in the repository.
Tool descriptors¶
To each available tool in our framework, we have a tool descriptor, i.e., a JSON file responsible for supplying all the information needed about the tool, such as the memory needed to execute it, the commands and the arguments of each command. The format of this file is given by the JSON schema presented in Listing 3.2.
{
"$schema": "http://json-schema.org/draft-04/schema#",
"type": "object",
"properties": {
"name": { "type": "string" },
"author": {"type": "string"},
"version": { "type": "string"},
"description": {"type": "string"},
"documentation": {
"type": "array",
"items": { "type": "string" }
},
"setup": {
"type": "array",
"items": { "type": "string" }
},
"toolType": {
"type": "string",
"enum": ["Unit", "splitting", "joinning", "listing"]
},
"requiredMemory": { "type": "integer" },
"recommendedCpus": { "type": "integer" },
"recommendedDiskSpace": { "type": "integer" },
"commands": {
"type": "array",
"items": {
"type": "object",
"properties": {
"name": { "type": "string" },
"command": {"type": "string"},
"description": {"type": "string" },
"priority": { "type": "integer" },
"argumentsComposer": { "type": "string" },
"arguments": {
"type": "array",
"items": {
"type": "object",
"properties": {
"name": {"type": "string" },
"argumentType": {"type": {
"enum": ["int","file","string","double","directory"]}},
"isRequired": {"type": { "enum": ["true","false"]} },
"description": { "type": "string" }
},
"required": ["name", "argumentType", "isRequired","description"]
}
},
"outputs": {
"type": "array",
"items":{
"type": "object",
"properties": {
"name": {"type": "string"},
"description": {"type": "string"},
"outputType": {"type": {
"enum": ["directory_dependent","file_dependent","independent"]}},
"argument_name": {"type": "string"},
"value": { "type": "string" }
},
"required": ["name","description","outputType", "argument_name","value"]
}
}
"inputs": {
"type": "array",
"items": {
"type": "object",
"properties": {
"name": { "type": "string" },
"description": { "type": "string" },
"inputType": { "type": "string",
"enum": [ "directory_dependent", "file_dependent", "independent"]}},
"value": { "type": "string" }
"required": [ "name", "description", "inputType", "argument_name", "value"]}}
},
"required": ["name","command","description","priority",
"argumentsComposer", "arguments","outputs", "inputs"]
}
},
"required": ["name","author","version","description",
"documentation","setup", "tool type",
"requiredMemory", "recomendedDiskSpace",
"recommendedCpus, "commands"]
}
}
Listing 3.2: JSON schema for specifying each tool included in the repository.
As an example, please see the tools descriptors that we have included in our tools’ repository example, such as the Velvet descriptor and the Trimommatic descriptor for Velvet and Trimmomatic tools, respectively. In our repository support library, each tool descriptor must be defined in a file named as Descriptor.json.
As defined on the previous JSON schema, a tool description must include its name, author, version, description, documentation, setup, toolType, required memory , recommendedCpus, recommendedDiskSpace and commands. The version property describes the version of the executable that is being considered by this descriptor. The documentation property allows to add a collection of links that contains documentation about the tool. The setup property contains all the scripts that must be executed before executing any command within the tool. For instance, for executing the Trimmomatic command, it must be previously installed the Java Runtime Environment. Thus, in the Trimmomatic descriptor, we include the setup presented in Example 3.1.
"setup" : [ "apt-get install -y default-jre" ]
Example 3.1: Trimmomatic command setup.
Command descriptions¶
commands is an array of JSON objects that describes each command within a tool. For instance, the Trimmomatic tool has only one command, but the Velvet tool has two commands, namely, velvetg and velveth.
For each command in the array commands it must exist its name, the command itself, its description, its priority, its arguments, the argumentComposer and its outputs. The priority of each command within a tool is important for defining execution dependency among commands within the same tool. For instance, in the Velvet tool, although not explicitly defined as an argument, velveth uses files produced by velvetg. If the files are already produced, then it is not necessary to execute velvetg if data is the same. Homever, if data differs from the last execution or is not yet produced, it must be assured that velvetg is executed before velveth. Therefore, we have added the priority property to each command to assign an integer that reflect the execution order within commands of the same tool which do not have it explicitly, but which is needed. The argumentsComposer item is the responsible for knowing how to concatenate the arguments, namely if arguments are passed as argName=argValue OR argName:argValue OR argName-argValue.
The many argumentComposer types supported by the NGSPipes repository support library are detailed in sub-section ArgumentsComposer. The arguments and the outputs are both arrays of JSON objects.
Argument descriptions¶
arguments is an array of JSON objects that describes each argument of a specific command. For each argument is required to define its name, its argumentType, if it is isRequired and its description. The type of each argument must be one of the following: integer number (int); file (file); text (string); real number (double) or a directory (directory). The isRequired property, which can be defined as true or false, indicates if is necessary to set a value to this argument or is an optional argument. As an example, consider the trimmomatic tool, which only has a command. For SINGLE END data, one output and input file are specified. Therefore, it is necessary to add to its descriptor the information specified in Example 3.2.
{
"name" : "outputFile",
"argumentType" : "file",
"isRequired" : "false",
"description" : "Specifies the name of output file."
},
{
"name" : "inputFile",
"argumentType" : "file",
"isRequired" : "false",
"description" : "Specifies the path to the fastq input file."
},
Example 3.2: arguments for Trimmomatic command
Both of the previous examples have the isRequired property set to false since for non SINGLE END data, trimmomatic execution uses pairs of input and output files, which are described in the tool descriptor by other arguments.
Output descriptions¶
output is an array of JSON objects that describes the outputs of each command. For each output is required to define its name, outputType, description,argument_name and value. Notice that the name passed as an argument to a command is not the name that is necessary to specify as a JSON property of the output JSON object. The name property refers to the name of the JSON object, not to the name of the file that is produced by the execution of a given command. Depending on the command, the name of the file that is produced by a given command can be set as an argument by the user or be an internal decision of the executing command.
Therefore, the independent outputType is used when an output value is specified inside of command and isn’t affected by any argument.
In this case, the value property of the JSON output object is set with the name that is internally generated by the corresponding command. An example of the output in descriptor file is depicted in Example 3.3.
{
"name" : "output",
"description" : "",
"outputType" : "independent",
"argument_name" : "",
"value" : "output.txt"
}
Example 3.3: Example of an output descriptor.
In the previous case, the argument_name is the empty string since there is no corresponding argument defined in the tool descriptor to set the name of the produced output file.
The outputType can also be file_dependent or directory_dependent.
An outputType is file_dependent if its value is specified in an argument and there is no specific directory that is created for keeping the generated output file. As an example, and taking into account the previous example of trimmomatic for SINGLE END data, the output is described in the tool description as presented in Example 3.4.
{
"name" : "outputFile",
"description" : "",
"outputType" : "file_dependent",
"value" : "",
"argument_name" : "outputFile"
},
Example 3.4: Example of an output descriptor.
In this case, the value property is set to the empty string since the name of the output file is specified by the user.
Moreover, the argument_name property defines the name of the JSON object that corresponds to the JSON object that defines the argument used for the specified the output file name.
The other type of output is
directory_dependent, which is used when an output value is added to a specified directory that is generated within the command execution.
In this case, the name of the directory is passed as an argument, but the name of the produced files are not passed as arguments. Instead, they are generated internally, within execution.
As an example, consider the velvet tool, where the commands outputs are of this type because they will be written to a directory, the first argument of velvetg and velveth, when executing both commands.
Therefore, since the output directory is a command argument, we have to specify in the tool descriptor a corresponding argument description, such as the one depicted in Example 3.5.
{ "name" : "output_directory",
"outputType" : "directory",
"isRequired" : "true",
"description" : "Directory where will be output files"
}
Example 3.5: Argument description in the case of a directory type
And thus, Example 3.6 illustrates of the output descriptor in the descriptor file, corresponding to the previous argument will be like:
{
"name" : "stats",
"description" : "",
"argumentType" : "directory_dependent",
"argument_name" : "output_directory",
"value" : "stats.txt"
}
Example 3.6: Output description when is dependent of an argument with type directory
Notice that the file name stats.txt is not passed as an argument to velveth nor to velvetg. Instead, it is generated internally and is stored in the output directory whose name was passed as an argument.
Input descriptions¶
input is an array of JSON objects that describes the inputs of each command. They are similar to Output descriptions. They help inferring dependencies between pipeline tasks.
ArgumentsComposer¶
In this subsection is listed all the argumentsComposer that are already included in our repository support library. The existing argumentsComposer are (name of the argumentComposer-> [corresponding format]):
- dummy -> []
- valuesSeparatedBySpace -> [value value value]
- nameValuesSeparatedByEqual -> [name=value name=value name=value]
- nameValuesSeparatedByColon -> [name:value name:value name:value]
- nameValuesSeparatedByHyphen -> [name-value name-value name-value]
- nameValuesSeparatedBySpace -> [name value name value name value]
- valuesSeparatedByColon -> [value:value:value]
- valuesSeparatedByVerticalBar -> [value|value|value]
- valuesSeparatedByHyphen -> [value-value-value]
- valuesSeparatedBySlash -> [value/value/value]
- valuesSeparatedByComma -> [value,value,value]
- trimmomatic -> [TRIMMOMATIC STYLE ArgCategory:arg:arg:arg]
- velvetG -> [VELVETG STYLE all arguments has format [name value] except output_directory that has format [value]]
Listing 3.3: Some argumentsComposer included in the repository support library.
Examples of the mapping of the arguments and output descriptions to command parameters.¶
As we can see in the Velvet tool manual, a simple execution of the velvetg command in the command line (without the NGSPipes System) after producing the executable with the make command is described in Example 3.7.
./velvetg velvetDir -cov_cutoff 5
Example 3.7: Executing velvetg command on the command line.
Therefore, the description of velvetg command, within the descriptor of velvet tool, must include two arguments descriptions, namely, one for the directory argument and other for the option _cov_cutoff. As we can observe in velvet descriptor file (https://github.com/ngspipes/tools/blob/master/Velvet/Descriptor.json), the JSON object for defining the arguments of velvetg command starts with the definitions depicted in Example 3.8.
{
"arguments" : [
{
"name" : "output_directory",
"argumentType" : "directory",
"isRequired" : "true",
"description" : "Directory where will be output files"
},
{
"name" : "-cov_cutoff",
"argumentType" : "float",
"isRequired" : "false",
"description" : "remove coverage nodes
AFTER tour bus or allow the system to infer it (default no removal)"
},
Example 3.8: Some velvetg arguments descriptions.
And, since the output directory produces output files, the produced output is directory_dependent as we can see in section “Output descriptions” within this section, the JSON object for defining the outputs of velvetg command starts with the descriptions depicted in Example 3.9.
"outputs" : [
{
"name" : "stats",
"description" : "",
"outputType" : "directory_dependent",
"argument_name" : "output_directory",
"value" : "stats.txt"
},
{
"name" : "preGraph",
"description" : "",
"outputType" : "directory_dependent",
"argument_name" : "output_directory",
"value" : "PreGraph"
},
Example 3.9: Some output descriptions for velvetg.
The values of these arguments ( velvetDir and 5, respectively) will be set in the pipeline specification. For more information about the pipeline specification, please consult (https://github.com/ngspipes/dsl/wiki).
Another example referred in this documentation is the Trimmomatic tool. As we can see in the Trimmomatic manual, For single-ended data, one input and one output file are specified. The required processing steps (trimming, cropping, adapter clipping etc.) are specified as additional arguments after the input/output files. Thus, it appears in description presented in Example 3.10 how to execute this command.
java -jar <path to trimmomatic jar> SE
[-threads <threads>] [-phred33 | -phred64] [-trimlog <logFile>]
<input> <output> <step 1> ...
Example 3.10: Executing Trimmomatic in the command line for single-ended data.
For paired-end data, two input files, and 4 output files are specified, 2 for the ‘paired’ output where both reads survived the processing, and 2 for corresponding ‘unpaired’ output where a read survived, but the partner read did not. Thus, it appears in the description presented in Exampl 3.11 how to executed this command in this version.
java -jar <path to trimmomatic.jar> PE
[-threads <threads] [-phred33 | -phred64] [-trimlog <logFile>] >]
[-basein <inputBase> | <input 1> <input 2>]
[-baseout <outputBase> | <unpaired output 1>
<paired output 2> <unpaired output 2> <step 1> ...
Example 3.11: Executing Trimmomatic in the command line for paired-ended data.
Thus, considering the SINGLE END DATA, a possible execution in the command line could be like the following
java -jar local/trimmomatic/trimmomatic-0.33.jar SE -phred33 ERR406040.fastq
ERR406040.filtered.fastq ILLUMINACLIP:local/trimmomatic/adapters/TruSeq3-SE.fa:2:30:10
LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36
**Example 3.12: **
In this case the input file is ERR406040.fastq and the output file is ERR406040.filtered.fastq. Thus, in the Trimmomatic tool description, we have included as arguments descriptions the ones described in Example 3.13.
{
"name" : "inputFile",
"argumentType" : "file",
"isRequired" : "false",
"description" : "Specifies the path to the fastq input file."
},
{
"name" : "outputFile",
"argumentType" : "file",
"isRequired" : "false",
"description" : "Specifies the name of output file."
},
{
"name" : "paired input 1",
"argumentType" : "file",
"isRequired" : "false",
"description" : "Specifies the path to the input file 1 of paired mode."
},
{
"name" : "paired input 2",
"argumentType" : "file",
"isRequired" : "false",
"description" : "Specifies the path to the input file 2 of paired mode."
},
Example 3.13: Trimmonatic arguments.
In the case of Trimmomatic command (please notice that Trimmomatic tool has only one command, with the same name), since both arguments inputFile and outputFile are only required in the SINGLE END data, their property isRequired was set to false.
With respect to the outputs, the Trimmomatic command description has the outputs described as in Example 3.14.
{
"name" : "outputFile",
"description" : "",
"argumentType" : "file_dependent",
"value" : "",
"argument_name" : "outputFile"
},
{
"name" : "paired output 1",
"description" : "",
"argumentType" : "file_dependent",
"value" : "",
"argument_name" : "paired output 1"
},
{
"name" : "unpaired output 1",
"description" : "",
"argumentType" : "file_dependent",
"value" : "",
"argument_name" : "unpaired output 1"
},
{
"name" : "paired output 2",
"description" : "",
"argumentType" : "file_dependent",
"value" : "",
"argument_name" : "paired output 2"
},
{
"name" : "unpaired output 2",
"description" : "",
"argumentType" : "file_dependent",
"value" : "",
"argument_name" : "unpaired output 2"
}
Example 3.14: Outputs description of Trimmomatic command.
As mentioned before, in the arguments and outputs descriptions, the values to be set to the arguments are done in the pipeline specification, as can be seen in the example in https://github.com/ngspipes/dsl/wiki. Notice that the Trimmomatic outputs are all file_dependent which means that its value is also an argument and thus is set by the user in the pipeline specification.
Tool configurators¶
The repository must also include at least one configurator for each tool. A tool configurator is responsible for adding all the information needed to define the execution context for executing the tool and its respective commands. Each tool configurator for each tool is given as a JSON file. Thus, for knowing all the available configurators for a specific tool, it exists, for each tool, a JSON file that lists all the JSON files that correspond to tool configurators for that tool. In our repository example and thus in our support implementation, these files appear at the root of each tool directory (please, see the a tool directory example).
For instance, in Blast tool, it can be observed that there is only a tool configurator (https://github.com/ngspipes/tools/blob/master/Blast/configurators.json), and the same is given as (https://github.com/ngspipes/tools/blob/master/Blast/DockerConfig.json).
List of configurators of a tool¶
For each tool, the list of the tool configurators that are available in a given repository are described in a JSON format in a file designed by Configurators.json. The format of the file that lists all the tool configurators files is given by JSON schema defined in Listing 3.4.
"$schema": "http://json-schema.org/draft-04/schema#",
"type": "object",
"properties": {
"configuratorsFileName": {
"type": "array",
"items":{ "type": "string" }
},
"required": [ "configuratorsFileName" ]
}
}
Listing 3.4: JSON schema for declaring the filenames of the configurators for a given tool.
Tool Configurators¶
As depicted in the previous schema, the file Configurators.json includes all the name of the files that corresponds to possible configurators for a given tool. Thus, for each file name included in onfigurators.json it exists a corresponding JSON file with the specific configuration. In our repository example and thus in our support implementation, the files for each specific configuration appears at the root of each tool directory ( please, see the a tool directory example). The format of this file is given by the JSON schema defined in Listing 3.5.
{
"$schema": "http://json-schema.org/draft-04/schema#",
"type": "object",
"properties": {
"name": {"type": "string" },
"builder": {"type": "string" },
"uri": { "type": "string" },
"setup": {
"type": "array",
"items": { "type": "string" }
}
},
"required": [ "name","uri","setup" ]
}
Listing 3.5: JSON schema for declarung each tool configurator.
Thus, a tool configuration is a JSON file with the following information: name of the file where is defined the execution context execution context (ex: DockerConfig); builder name of the execution context (ex: Docker);
setup}, i.e., the scripts that are necessary to execute to assure the existence of the execution context; and the uri where the tool is. Next example describes that the tool is on a docker image and thus is necessary to install docker in the execution context.
{
"name" : "DockerConfig",
"builder": "Docker"
"uri" : "simonalpha/ncbi-blast-docker",
"setup" : [
"wget -qO- https://get.docker.com/ | sh"
]
}
Example 3.15: Example of a tool configurator for the blast tool.
Defining your own tool repository¶
Each user can define its own tool repository, locally or remotely and use NGSPipes support library. The simplest way to do this it to use an hierarchical directory system approach, either locally or remotely. For a different form of structuring data, it would probably be necessary to extend NGSPipes support library.
Using an hierarchical directory system approach¶
In this section it will be described how to use an hierarchical directory system approach for defining a new tool repository, locally and remotely. For the remote case, we will use github as an example. In both cases, it is necessary to create a directory for each tool. The directory name will be seen as the tool name (the tool identifier on the repository) and is exactly the same name that is used in the pipeline definition and in the file Tools.json.
In the file Tools.json there will be a tool name for each available tool in the repository.
Each tool directory keep all the information about that tool, namely its description, its logotype, its configurators and the file name of its configurators. As mentioned before, the tool descriptor, which includes all the metadata needed to describe a tool, is given in a JSON file. As a convention, each tool descriptor file name is Descriptor.json. With respect to the logo file it should be a png file named as Logo.png. The logo file is optional. The file where is kept the file name of the configurators for a tool is also given as a JSON file, always designed as Configurators.json. For each file name specified in this file, there must exist the respective configurator JSON file.
Define a new repository locally¶
For defining a new repository in our own computer we have first to create a directory that will be our tool repository (ex: named as tools). Then, add to tools directory the file Tools.json and for each tool name that appears in this file, which identify a specific tool, create a new directory in tools. Each new created directory inside tools must have the corresponding name used in Tools.json to identify the tool. Each tool directory must contain the data described in the beginning of subsection “Defining your own tool repository”.
Define a new repository on github¶
After log-in in github, create a new repository (ex: named as tools). The endpoint of this new repository will the tool repository.
Then, after cloning your repository to your computer, it will appear a directory named as tools. Then, do the same steps of a section “Define a new repository locally” within this subsection. After that, synchronize the repository.
Tool Types¶
For supporting data partitioning in the engine for cloud, which will allow to executing in multiple machines partitions of data at the same time and thus increase process efficiency, the ´tool type´should be specified in a tool descriptor. Notice that this feature has only impact in the engine for cloud solution. There are four different tool types:
- data processing tools, i.e., unit;
- listing tools, i.e., splitting;
- splitting tools i.e., joining;
- joining tools i.e., listing.
Unlike unit processing tools, where each command is mapped into one task, a splitting command within a splitting tool generates one task corresponding to the splitting of the file plus N tasks per command, where N is the number of partitions of the file whereas each task processes a partition of the file. Data partitioning allows users to work with and process multiple files having to specify each command only once, while treating the files like a single file. It means that when users split a file in ten, for instance, they do not have to include the same tool ten times in the pipeline for every partition: the analyser will do that for them.
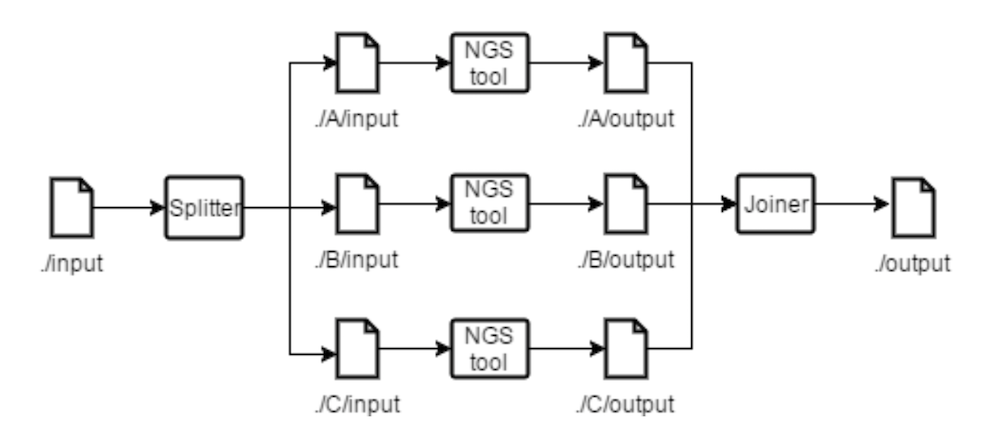
Figure 3.1 Splitting and Joining tools example.
Figure 3.1 shows how a file named input is split originating three different files with the same name, stored in directories with different names. For each partition the analyser will generate a directory where it stores the file partition with the same name it had before being partitioned. For every command specified in the pipeline description that uses the partitioned file, it is generated a task where the input path (partitioned file’s path) is concatenated with the name of the directory where the partition of the file is stored. Multiple directories are created to avoid name collision between files generated. Joining tools generate a task to join the partitions with the name of the input, that are stored in analyser generated directories (through either splitting or listing) corresponding to that input. Commands whose input depend on the join output will no longer have their tasks multiplied per partition. In Figure 3.2 it is depicted an example where a user wants to process different files of the same type, using the same tools, without having to specify each command more than once. Listing tools move and rename the files to match the same pattern as the splitting tools. After files are listed, they can be treated as one, as if it had been a split. While the splitting tools are used to partition data files and apply the same command or set of commands to each partition, the listing tools allow users to
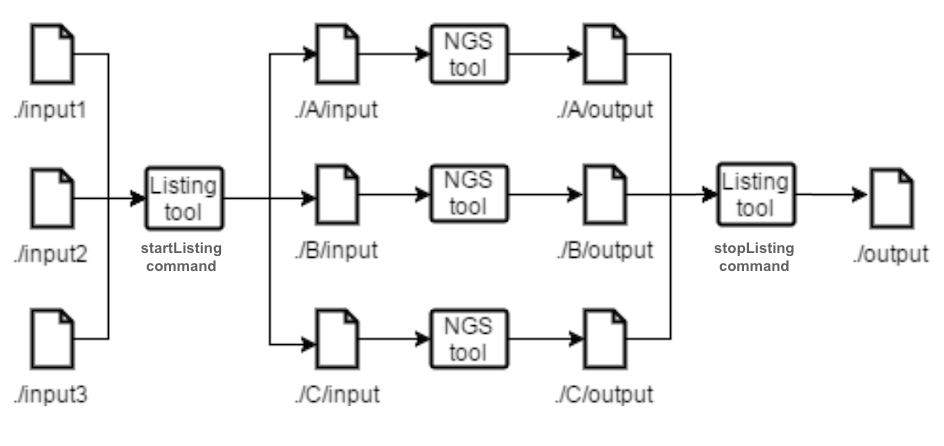 Figure 3.2 Listing and Joining tools example.
Figure 3.2 Listing and Joining tools example.
apply the same command or set of commands to a list of specified files. Listing tools generate a task to move the files to the newly generated directories and change the files names to match the name used in the .pipes file. Listing tools generate the same tasks as splitting and joinning tools on commands that depend from them. The listing tools purpose is to allow a user to provide multiple files and treat them as one in the .pipes file. The listing tools have two commands, startlisting and stoplisting that are similar to split and join tools, respectively, but applied when the input are multiple files (passed as a zip file).
The way data partitioning and dependencies inference is implemented, allows users to benefit from parallelization without adding complexity to the DSL and the process of writing a .pipes file.
The analyser also skims all the outputs that will be produced in order to create a list of the directories that have to exist for the swift and correct execution of the pipeline. These directories are stored in an array on the intermediate representation, under the directories property.
With the current version of the analyser, we have achieved a solution that allows users to split data and that allows to infer a topological graph from task dependencies, enabling the parallelization of the pipeline execution, without increasing the DSL complexity.
tool "Listing" "DockerConfig" {
command "startListing" {
argument "referenceName" "reads.sam"
argument "filesList" "snp-pipeline-master/snppipeline/data/lambdaVirusInputs/reads1.sam snp-pipeline-master/snppipeline/data/lambdaVirusInputs/reads2.sam snp-pipeline-master/snppipeline/data/lambdaVirusInputs/reads3.sam snp-pipeline-master/snppipeline/data/lambdaVirusInputs/r
}
}
Listing 3.6 An example of tool with type listing type.
More details on the type of tools can be found in this report.